journey to tocky via cca
Transitioning from Molecular Immunology to Systems Immunology
Until 2007, I focused on the molecular biology of Treg, publishing seminal papers for the Foxp3-Runx1 interaction (Ono et al., 2007).

However, upon realising the fundamental problem with Treg, I chose an unconventional path - extending my expertise to computational biology💻, which would become pivotal in developing Tocky.
First Independent Studies in Kyoto
As I was promoted to Assistant Professsor in 2007, I secured grant support from the Kato Foundation in 2008 for my first transcriptome analysis using microarray 🧬.
However, the complexity of the data was overwhelming, challenging the simplistic analytical methods of the time.

The Lack of Appropriate Methods
Back then in 2006 - 2007, immunology papers mainly used simple methods like log2 fold change only, which was not convincing to me.
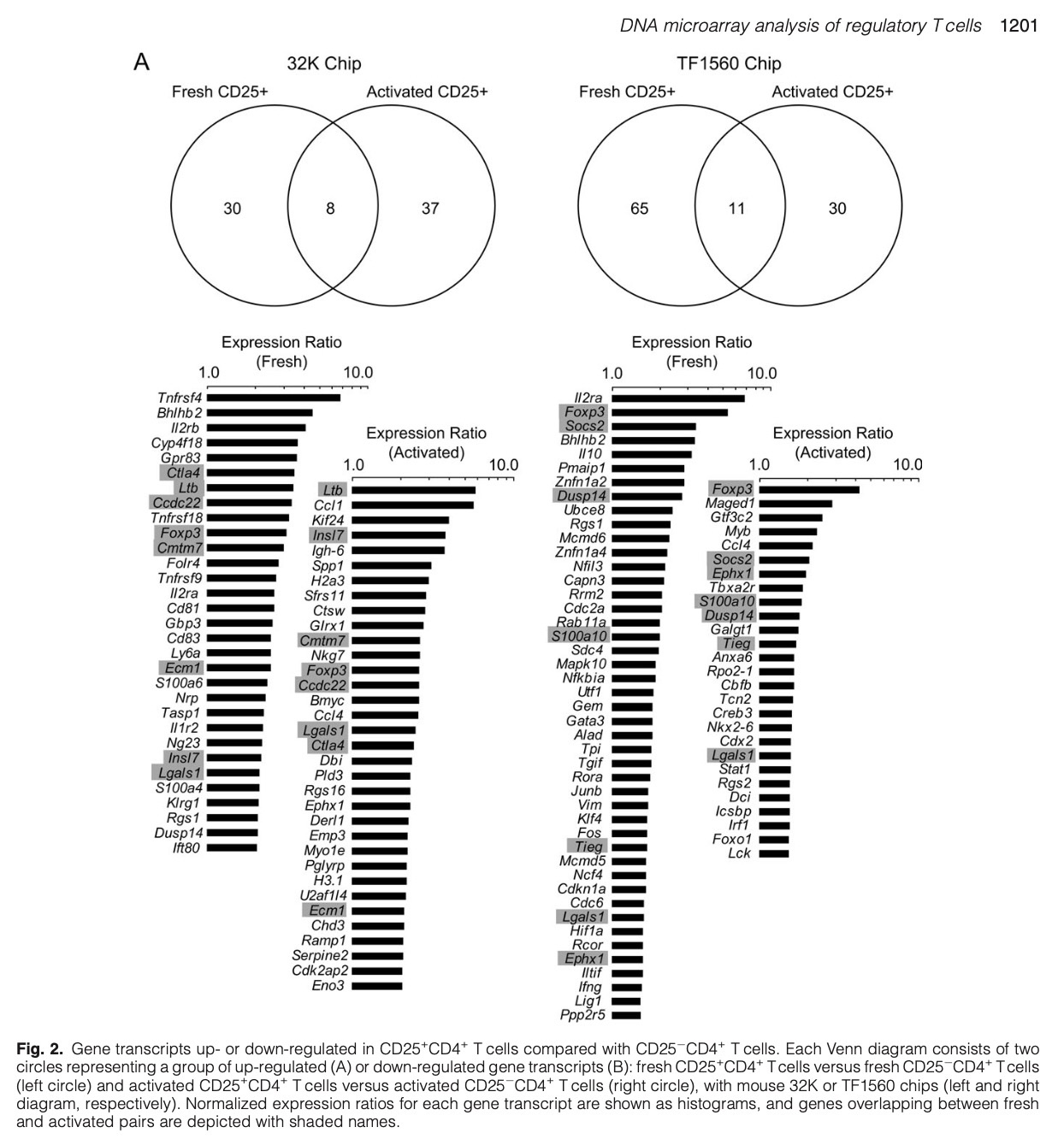
Exploration and Collaborations in Kyoto
Thus, in 2008, I aimed to develop more sophisticated methods for immunological genomic analysis, identifying Principal Component Analysis (PCA) as a promising candidate 🤔🔍.
In 2008, Dr Manabu Kano, an expert in PCA in Eng. Dept, Kyoto Univ, provided invaluable guidance, teaching me mathematical foundations of PCA in data analysis.
This collaboration was a pivotal step in my journey to redefine Treg in a data-oriented manner🎓💡.
In addition, Prof Toshio Sugiman, an expert in psychometrics🧠📈, introduced me to Hayashi’s Quantification Method III, a tool for quantifying group relationships in psychology.
Hayashi’s statistical philosophy profoundly inspired me, and this will lead to the development of the groundbreaking method five years later.
Extending Research Fields: From Kyoto to the UK.
Aiming to adapt Hayashi’s method and PCA for genomic data in immunology, I obtained an HFSP fellowship and thereby moved from Kyoto🇯🇵 to UCL🇬🇧 , eventually led to the development of Canonical Correspondence Analysis, originally by ter Braak, for genomics (Ono et al., 2013).
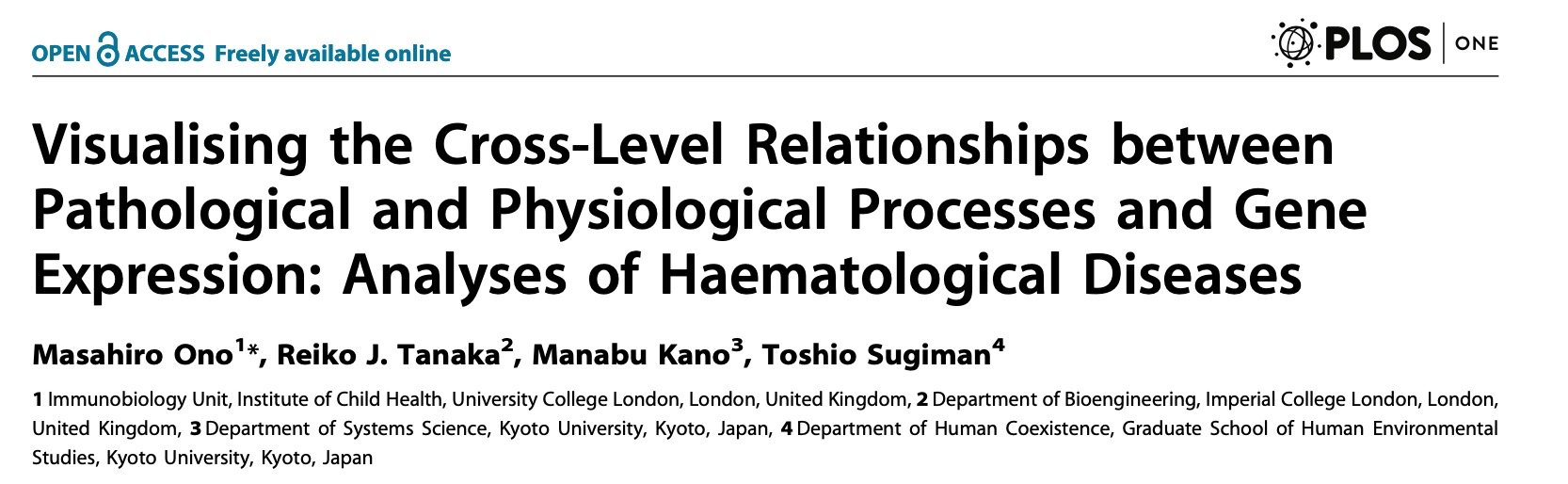
🔬Canonical Correspondence Analysis is an efficient method for [qunatitative anlaysis of cell phenotype and gene activities, focusing on certain biological processes]((https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-15-1028). In addition, it can classify cells using a machine learning approach (Ono et al., 2014)🧬💻.
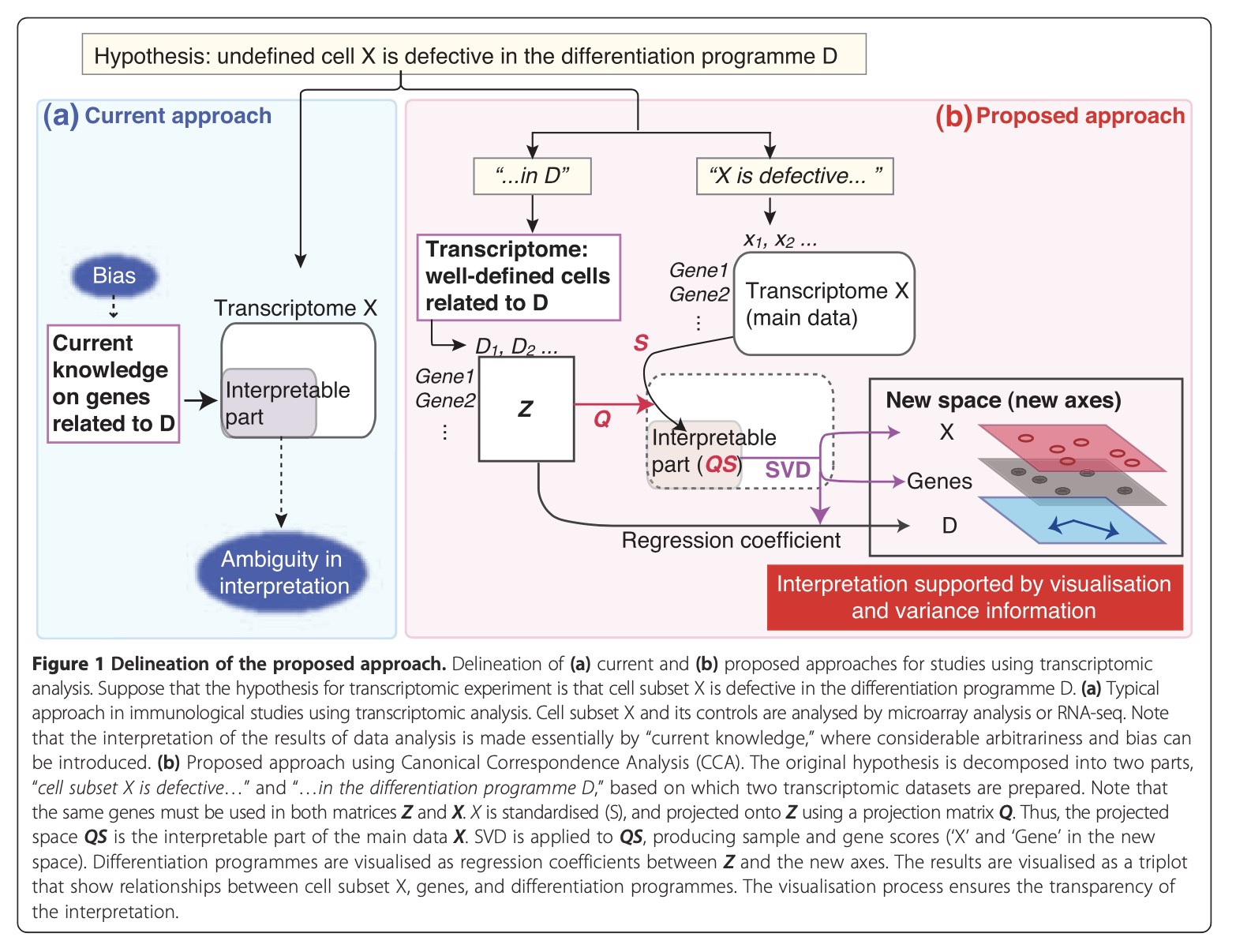
The Journey to Tocky is Through CCA
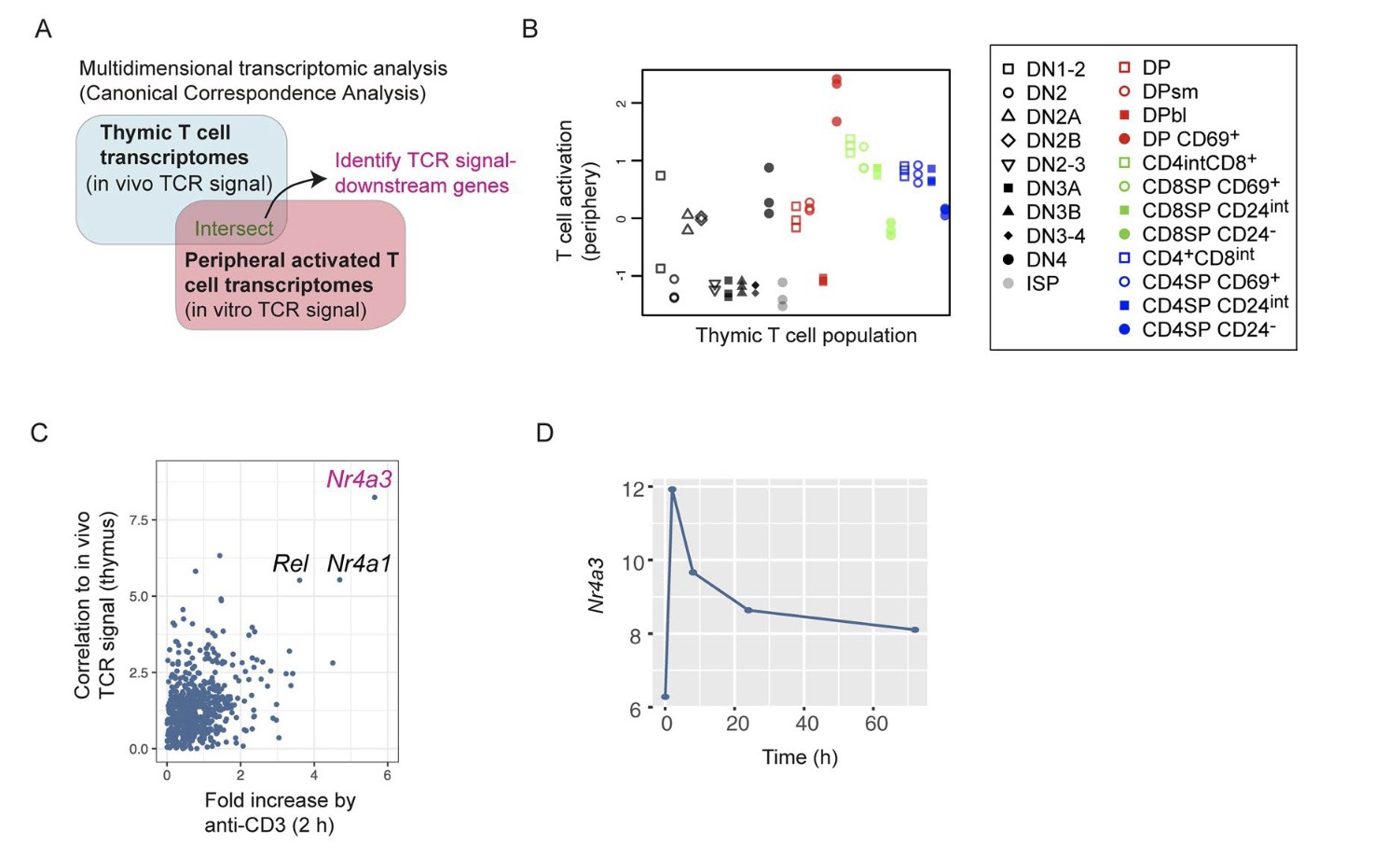
Importantly, CCA was utilized to identify Nr4a3 as the best correlate to TCR signalling, which was a key step in developing Nr4a3-Tocky (Bending et al., 2018) 🐭🔬.
References
2018
- A timer for analyzing temporally dynamic changes in transcription during differentiation in vivoJournal of Cell Biology, 2018The foundational publication introducing Tocky technology by the Ono lab, marking a breakthrough in T cell and B cell studies.
2014
- Visualisation of the T cell differentiation programme by Canonical Correspondence Analysis of transcriptomesBMC Genomics, 2014This study establishes CCA as a vital quantitative tool for transcriptome analysis in immunological datasets.
2013
- Visualising the cross-level relationships between pathological and physiological processes and gene expression: analyses of haematological diseasesPLoS One, 2013The pioneering study developing Canonical Correspondence Analysis (CCA) as a genomics method, establishing a novel approach for transcriptome analysis.
2007
- Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1Nature, 2007
Enjoy Reading This Article?
Here are some more articles you might like to read next: