Tocky's Debut
The original report of Tocky from the Ono lab
In 2013, upon receiving the prestigious BBSRC David Phillips Fellowship, Masahiro Ono launched his groundbreaking project to develop a novel methodology for analyzing the temporal dynamics of T-cell activities using a Fluorescent Timer protein—Timer-of-cell-kinetics-and-activity (Tocky).
This culminated in a seminal publication in 2018 (Bending et al., 2018).
Discovery of Nr4a3 as a Reliable Reporter Gene for Cognate Antigen Signaling
Upon establishing my laboratory at University College London in 2013, I embarked on developing an entirely new approach to comprehend T-cell dynamics in vivo.
Through Canonical Correspondence Analysis, Nr4a3 emerged as the gene with the highest correlation to T-cell receptor (TCR) signaled T-cells in both the thymus and periphery.
Crucially, Nr4a3 is not expressed in resting T-cells or B-cells. However, antigen signaling (specifically, cognate antigen signaling) triggers Nr4a3 expression in both T-cells and B-cells.
Development of Nr4a3-Timer Reporter Mice (Nr4a3-Tocky)
The initiation of the Timer method led to a dedicated project in my isolated lab—I pressed forward alone, without hiring any staff, to maximize time and resources for developing a new animal model.
The development of Nr4a3-Timer reporter mice, Nr4a3-Tocky, utilized the Bacterial Artificial Chromosome approach for several reasons, which will be detailed in a future post. My extensive experience with transgenic mouse strains in Japan facilitated the successful development of Nr4a3-Timer mice in 2013.
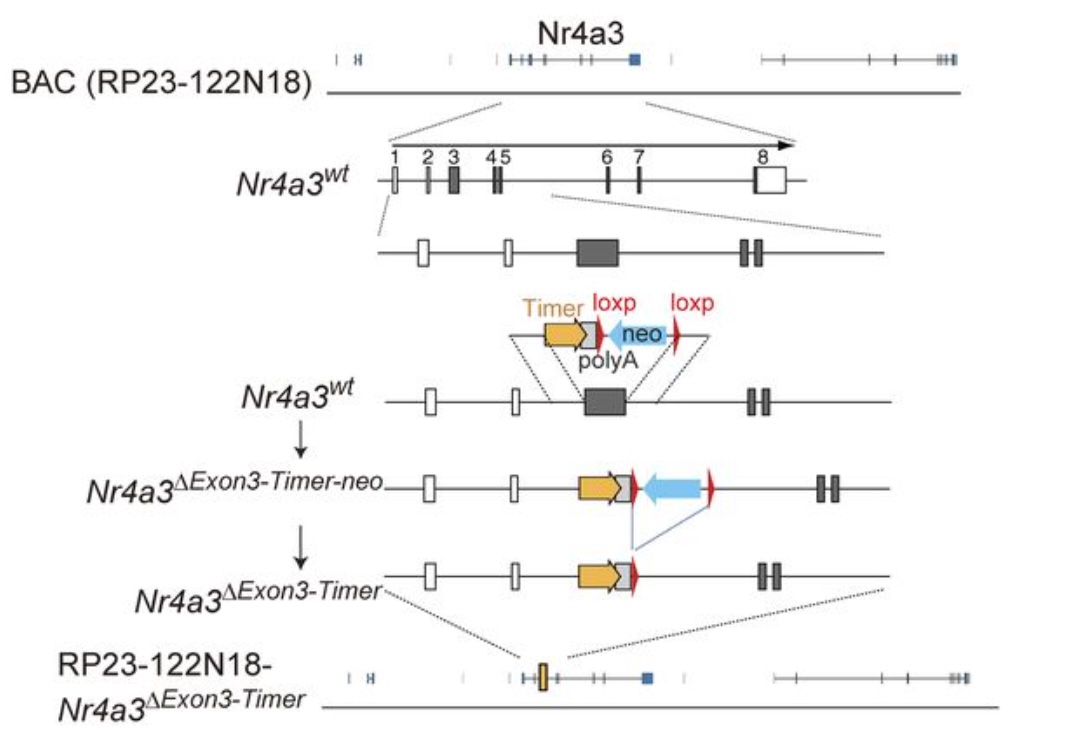
In 2014, I expanded the transgenic colony by hiring a research technician, followed by the recruitment of a post-doc in 2015.
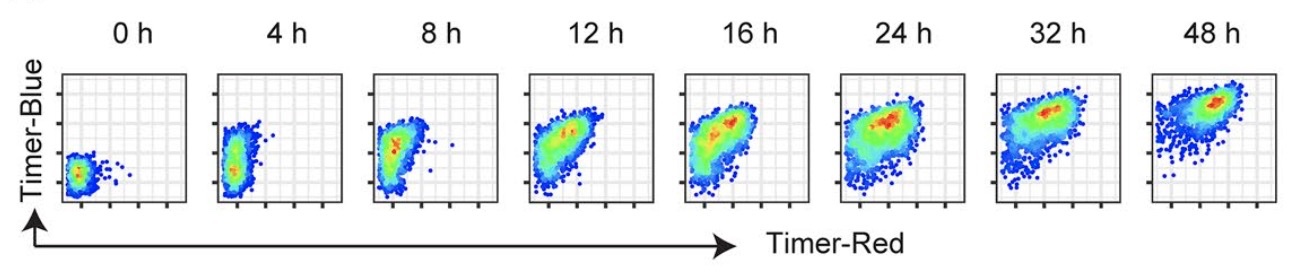
Upon TCR stimulation, Nr4a3-Tocky T cells exhibit initial timer blue fluorescence within the first hours, progressively maturing into red fluorescence as Timer proteins mature. Interestingly, persistent transcription drives the cells to align along a diagonal line between the Blue and Red axes.
Rejection and Resilience
Despite a smooth start, we encountered significant obstacles—neither editors nor reviewers were able to recognize the importance of the Timer system. Interestingly, many claimed that the Timer approach was “not novel.”
Now, I take pride in the fact that our paper was rejected nearly 20 times, which is a robust testimony for the true novelty of the Tocky approach.
I should accurately count the exact number and compile their comments for future reference.
Development of Novel Tocky Data Analysis Method
This challenge prompted my second innovation: developing a novel, data-oriented analysis method for analyzing Timer fluorescence data.
As demonstrated in the figure above, Timer fluorescence data are continuous, which highlights the limitations of traditional gating methods in fully leveraging the Timer’s capabilities. Recognizing the necessity for a tailored approach, I pioneered a novel trigonometric data transformation known as the “Timer Angle.” This method significantly enhances the analysis of T-cell dynamics. Details of this innovative approach will be explored in a future blog post.
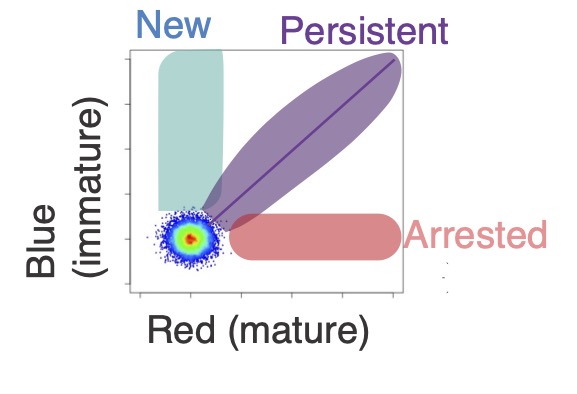
The Tocky technology, equipped with the quantitative data analysis methods and meticulously designed Timer reporter systems, provides unique solutions to immunology and beyond.
References
2018
- A timer for analyzing temporally dynamic changes in transcription during differentiation in vivoJournal of Cell Biology, 2018The foundational publication introducing Tocky technology by the Ono lab, marking a breakthrough in T cell and B cell studies.