temporal trajectories elucidated by tocky
research paper reporting tockyDevelopment
Deciphering Temporal Dynamics in Cell Development: Introducing Our Multidimensional Toolkit Using Tocky Technology
Date: 1 December 2024
Author: Masahiro Ono
I am thrilled to share groundbreaking developments from our latest research. Our study, titled “A multidimensional toolkit for elucidating temporal trajectories in cell development in vivo,” introduces an innovative computational tool that exploits the multidimensional power of our Tocky technology to decipher time-dependent processes in cellular development (Ono & Crompton, 2024).
The link to paper is here.
Background: Overcoming Challenges in Analyzing Temporal Dynamics of Developing Cells in Vivo
During cell development, progenitor cells respond to key signals by altering gene expression and phenotypes, initiating dynamic developmental processes. However, it is extremely challenging to capture these dynamic processes at the single-cell level in vivo, primarily due to the stochastic nature of signal transduction and the lack of adequate methodologies in both experimental and computational analysis.

The Innovation: Tocky Technology and Multidimensional Analysis for High-Dimensional Flow Cytometric Timer Data
At the core of our approach is the Tocky technology, a tool that uses a Fluorescent Timer reporter gene to track the time elapsed since a cell receives a developmental cue. This technology comprises experimental Timer reporter systems and computational algorithms to analyze Timer data.
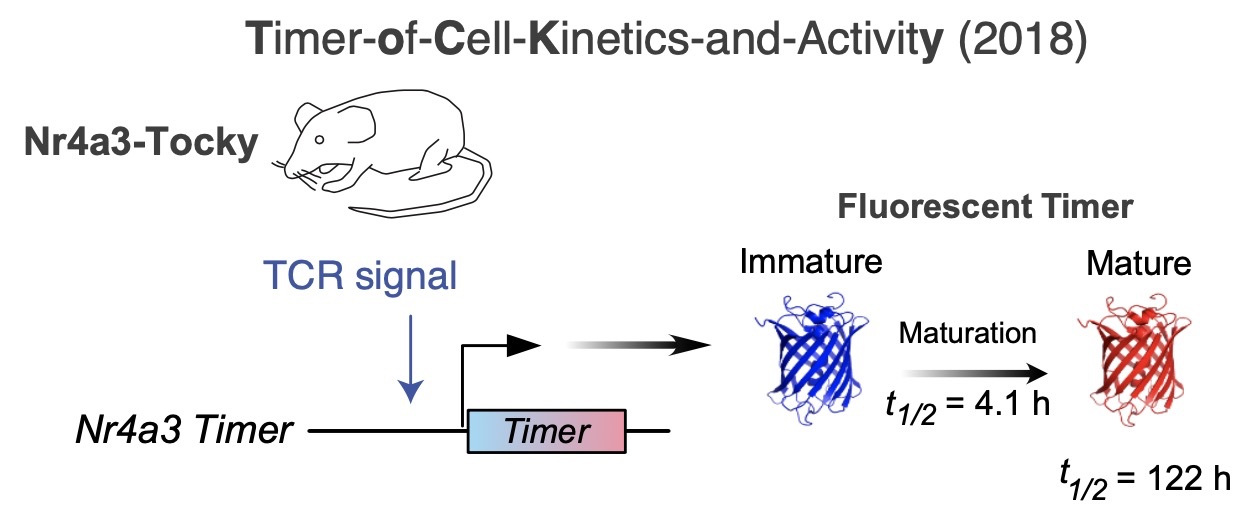
With advancements in cytometry technologies, the combined analysis of high-dimensional marker profiles and Timer fluorescence data can provide opportunities to fully integrate this temporal measurement with high-dimensional flow cytometric data. This approach allows for deciphering time-dependent changes during cell development, leveraging experimental data in a data-oriented manner.
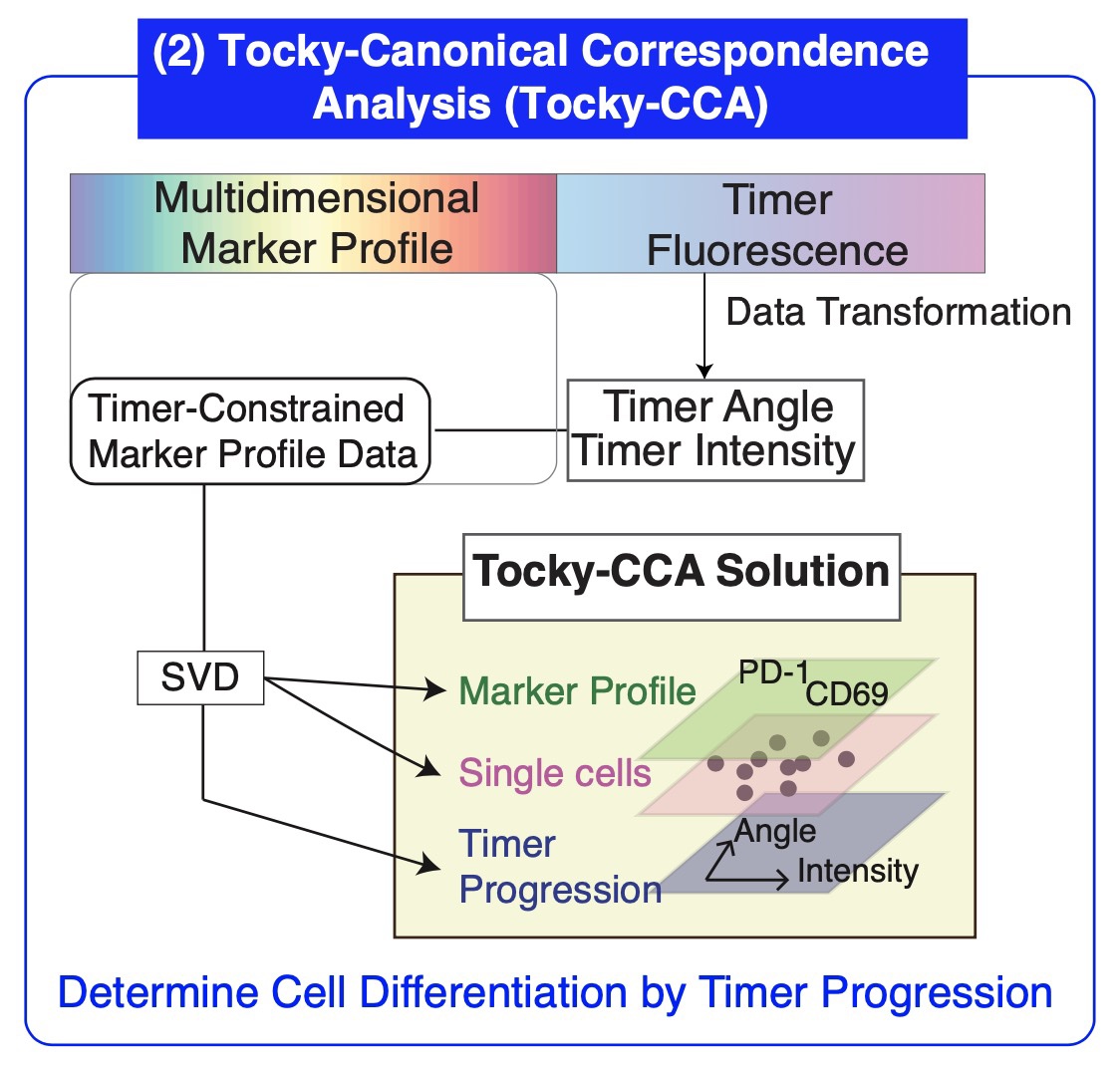
Solution: Canonical Correspondence Analysis (CCA) Meets Flow Cytometry
A pivotal advancement in our toolkit is the application of Canonical Correspondence Analysis (CCA), adapted to manage the complexities of flow cytometric data. CCA was originally developed by ecological statistician Cajol ter Braak. From 2009 to 2014, I recognized CCA’s potential in genomics and developed it as a method for genomic analysis.
📄 Read the blog entry for the development of CCA
Now, for the first time, CCA is adapted to high-dimensional flow cytometric analysis, elucidating the time-dependent changes as retained in Timer fluorescence. This adaptation allows for the cross-analysis of Timer progression with the multifaceted marker profiles of cells, thus elucidating temporal trajectories of cellular development through time.
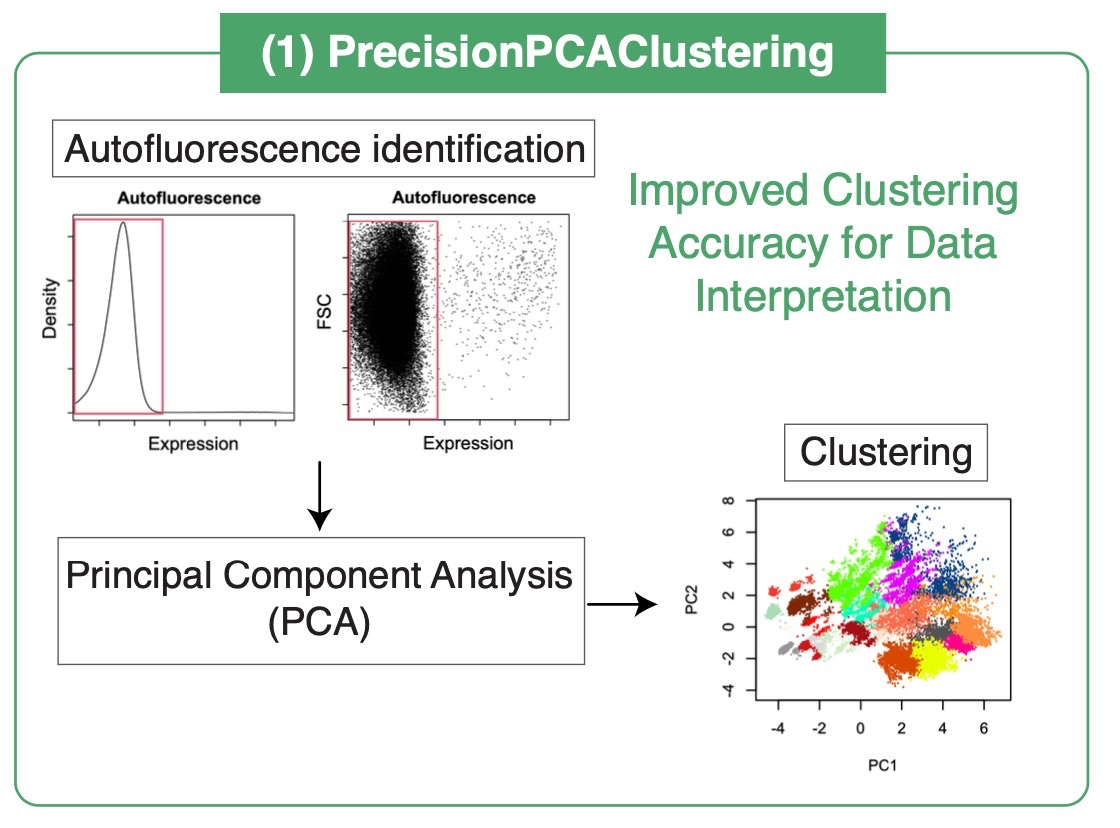
A Novel Principal Component Analysis-Based Approach to Overcoming Stochasticity in Clustering
Autofluorescence in flow cytometry can obscure true signals, complicating data analysis. Our innovative PrecisionPCAClustering algorithm tackles this by distinguishing genuine marker expressions from background noise. This enhancement not only improves the accuracy of clustering but also preserves the continuity of developmental pathways, which is crucial for detailed trajectory analysis.
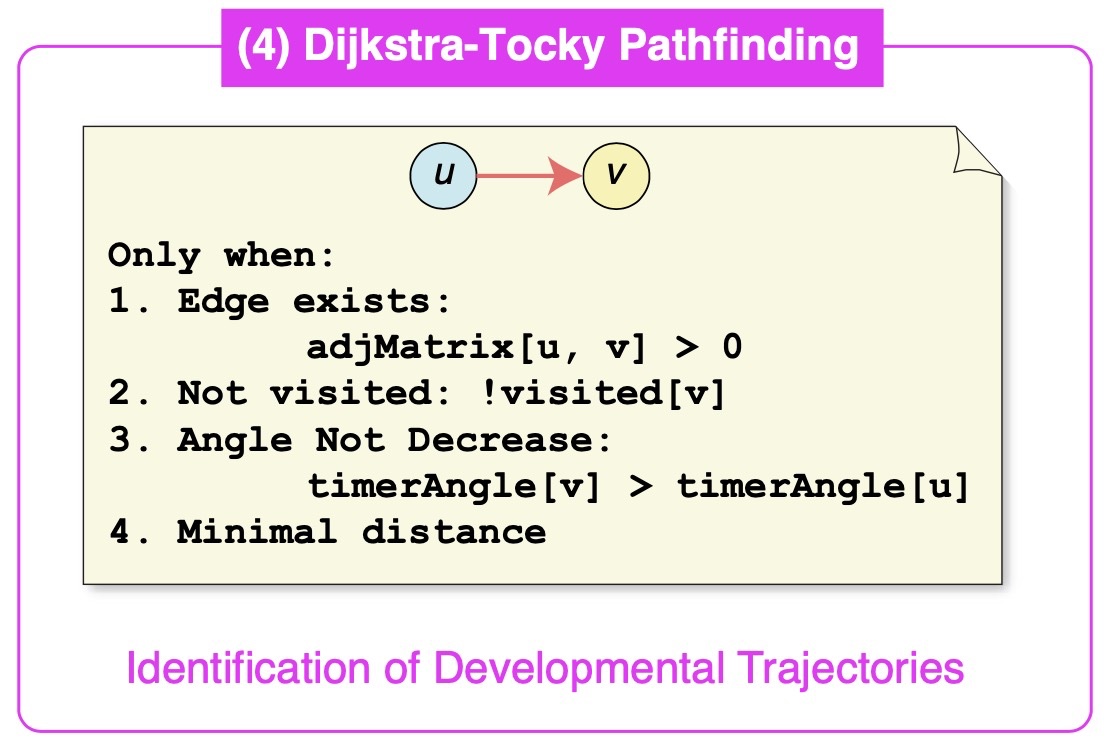
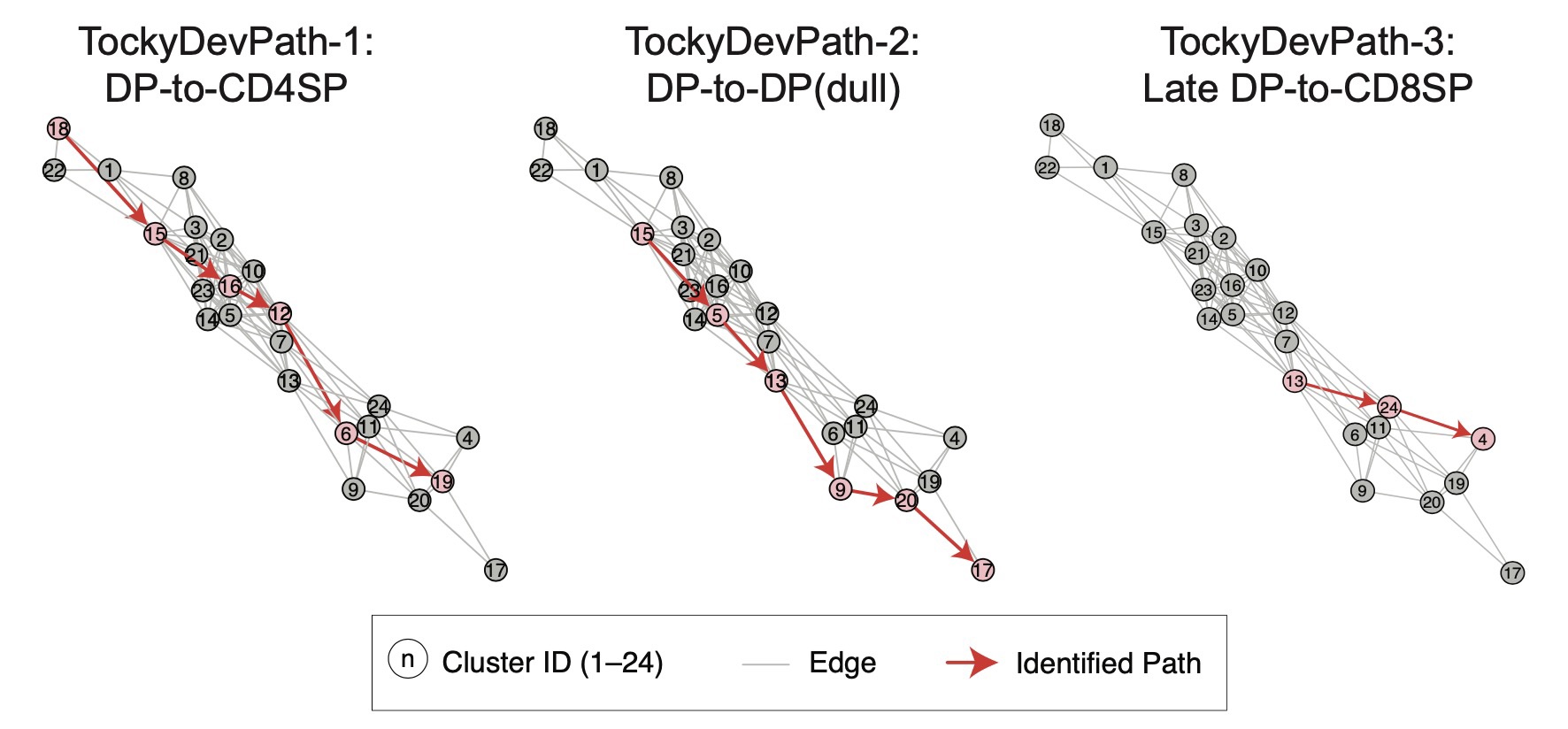
Unveiling Developmental Trajectories with Dijkstra-Tocky Pathfinding
To further elucidate developmental pathways, we developed the novel Dijkstra-Tocky pathfinding algorithm, an adaptation that uses time-encoded data to trace the most probable paths of cell differentiation, considering Timer progression and multidimensional marker profiles. This technique has helped us discover distinct trajectories for CD4 single-positive T-cells, providing unprecedented insights into their differentiation processes.
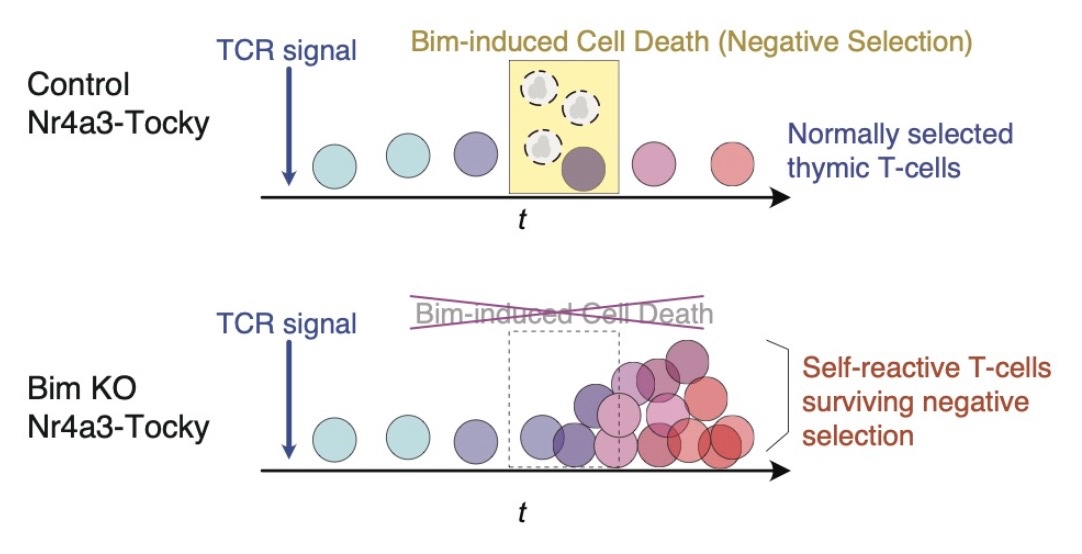
Case Study: Thymic T-Cell Development in Nr4a3-Tocky Mice
Nr4a3-Tocky mice
Our toolkit’s effectiveness is exemplified in a study of thymic T-cells from Nr4a3-Tocky mice. In this model, TCR signaling acts as a developmental trigger, influencing cell fate and maturation. By analyzing Timer progression in conjunction with multidimensional marker profiles, we decipher thymic T-cell development in an unprecedented manner using our novel multidimensional Timer approach.
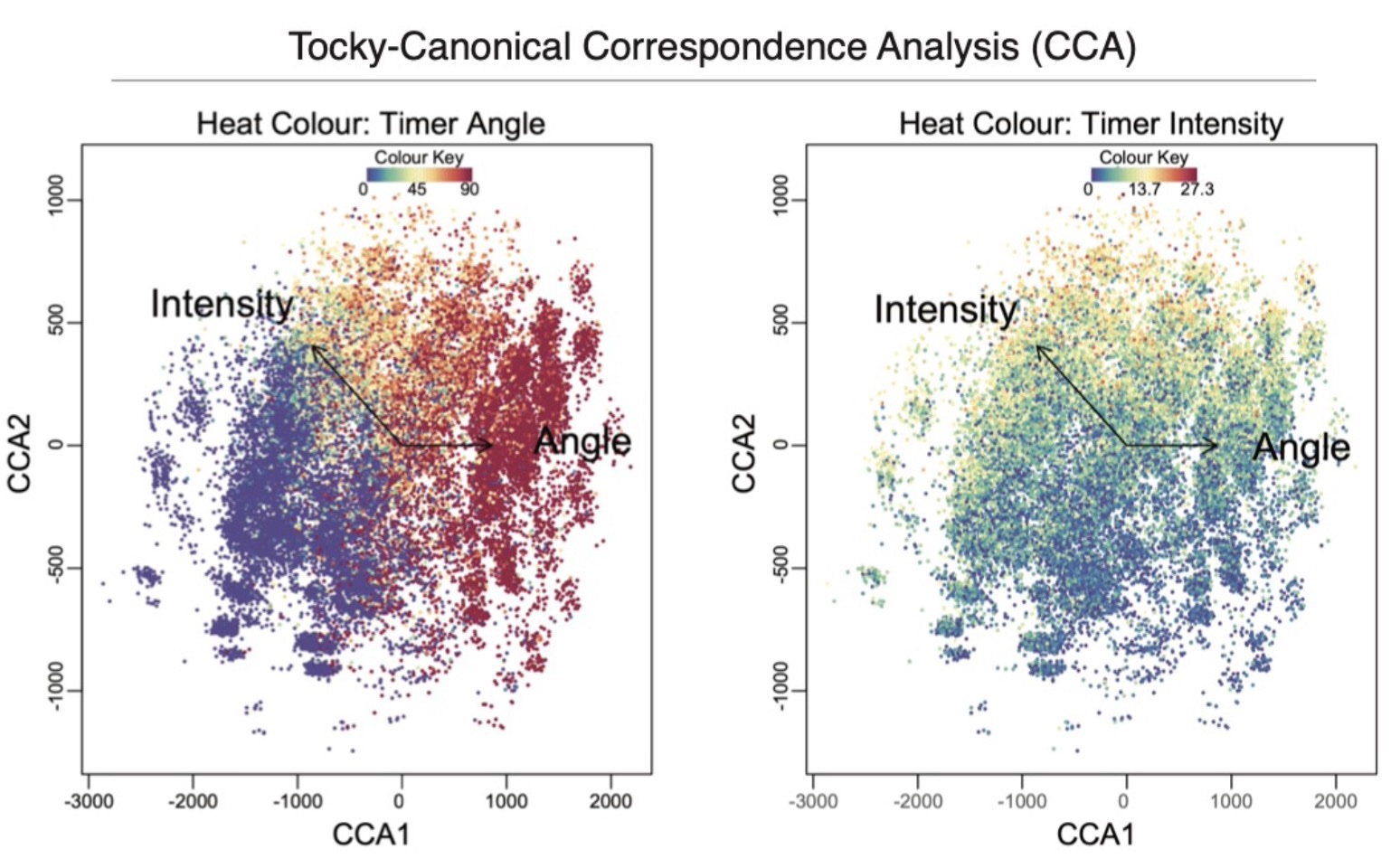
Bim KO Nr4a3-Tocky mice and Tocky-CCA Analysis
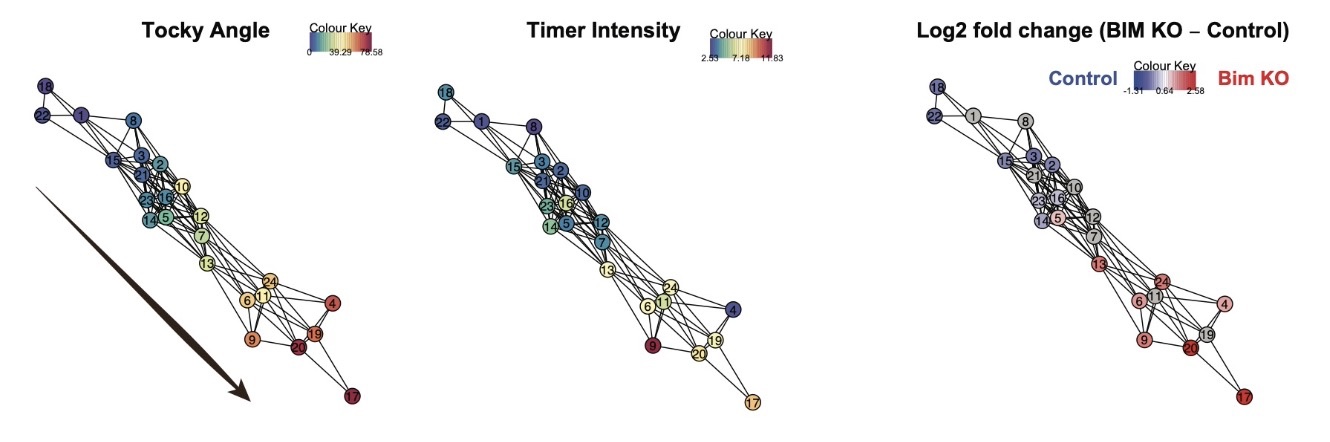
Furthermore, by examining high-dimensional flow cytometric data from Bim KO Nr4a3-Tocky mice—mice with the Bcl2l11 (Bim) gene knocked out in the Nr4a3-Tocky strain—we can highlight the Bim-dependent death of developing T-cells, marked by Timer maturation during negative selection.
Bim KO Nr4a3-Tocky toolkit elucidates the timing of cellular death during development in an experimental and data-oriented manner, marking a key milestone in developmental biology.
Tocky-CCA Network Analysis
Dijkstra-Tocky pathfinding
Dijkstra-Tocky pathfinding algorithm enables identification of time-dependent trajectories.
TockyDevelopment: A Multidimensional Toolkit for High-Dimensional Flow Cytometric Fluorescent Timer Analysis
The methodologies and computational algorithms are encapsulated in our multidimensional toolkit, TockyDevelopment, which not only enhances our understanding of T-cell development but also has broad applications across various cell types and biological processes. It represents a significant leap forward in single-cell analysis, enabling, for the first time, the experimental measurement of time-dependent trajectories in developing cells.
In conclusion, our research presents a comprehensive solution to the longstanding challenge of analyzing time-dependent cellular dynamics. By integrating innovative computational strategies with Tocky technology, we have opened new pathways for understanding and manipulating cellular development. We look forward to its adoption and further development by the broader scientific community, confident that it will spur numerous discoveries in developmental biology and beyond.

References
2024
- A multidimensional toolkit for elucidating temporal trajectories in cell development in vivoDevelopment, Nov 2024