TockyDevelopment
Multidimensional Tools for Flow Cytometric Fluorescent Timer Data
TockyDevelopment: Multidimensional Tools for Flow Cytometric Fluorescent Timer Data
Author: Dr. Masahiro Ono
Date: 24 November 2024
Link to the TockyDevelopment Documentation Page
Introduction: Fluorescent Timer as Experimental Tool and Development of the Tocky Concept
The potential of Fluorescent Timer proteins
Fluorescent Timer proteins uniquely alter their emission spectra over time, making them powerful tools for monitoring dynamic cellular processes. These proteins are pivotal for understanding the intricate temporal dynamics of cellular events. Despite their potential, analyzing Timer fluorescence data in flow cytometry is often hampered by variability in instrument settings and the lack of standardized data preprocessing methods.
Tocky: A Breakthrough in Fluorescent Timer Analysis
A significant advancement was made in 2018 when the Ono lab introduced Tocky, a novel concept for analyzing Fluorescent Timer data. This approach encompasses data normalization and transformation methods (see Introduction to the TockyPrep package) and quantitative analysis methods for Fluorescent Timer data (see Introduction to the TockyLocus package). However, a computational implementation of this methodology was not yet available.
The breakthrough in the anslysis of Fluorescent Timer data was the introduction of the polar coordinate, or trigonometric transformation of Timer fluorescence data, which produces the key variables: Timer Angle and Timer Intensity.
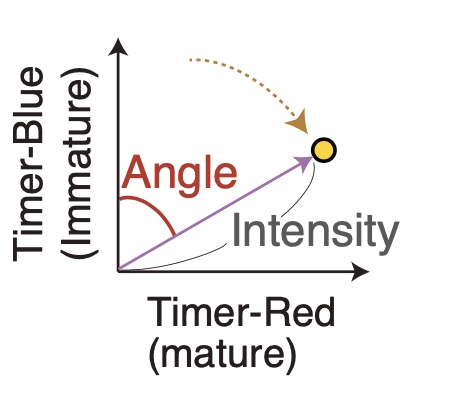
However, to effectively use the approach, a dedicated computational tool was required to normalize and transform Timer fluorescence data. To address these challenges, we have recently developed the TockyPrep package. This R package provides a comprehensive suite of tools designed to automate the preprocessing, normalization, and trigonometric transformation of Timer fluorescence data. TockyPrep specifically addresses the normalization of immature and mature Timer fluorescence as a critical step for robust downstream analysis.

TockyDevelopment: The R Package for Multidimensional Analysis of Flow Cytometric Fluorescent Timer Data

Aim of the TockyDevelopment Package
The aim of the TockyDevelopment package is to offer functions for analyzing the temporal dynamics of developing cells during development, utilizing Fluorescent Timer protein reporters, cross-analyzing Timer data and multidimensional marker expression profiles. The package enables identification of time-dependent trajectories using Fluorescent Timer data.
Key Features
The TockyDevelopment package offers functions for analyzing the temporal dynamics of developing cells during development, utilizing Fluorescent Timer protein reporters, cross-analyzing Timer data and multidimensional marker expression profiles. The package enables identification of time-dependent trajectories using Fluorescent Timer data.
-
Integrative Analysis with TockyCCA
- Implements TockyCCA, a variant of Canonical Correspondence Analysis (CCA) optimized for multidimensional flow cytometric data involving Fluorescent Timer proteins.
- Allows integrative analysis of marker expression profiles and Timer progression without centering the environmental variables (Timer Angle and Intensity), preserving their directional and magnitude properties.
-
Network Analysis of Cell Clusters
- Analyzes TockyCCA outputs using network analysis to elucidate relationships between cell clusters.
- Identifies potential developmental trajectories by examining the connectivity and transitions between clusters.
-
Pathfinding Analysis with Dijkstra-Tocky Algorithm
- Employs a novel pathfinding algorithm, Dijkstra-Tocky, to identify time-dependent trajectories based on network analysis outputs.
- Pinpoints developmental paths by finding the shortest paths in the network, representing the most probable sequences of cellular transitions.
Availability
- TockyDevelopment is available at GitHub: TockyDevelopment.
Package Documentation
The TockyDevelopment package documentation is available online:
This site includes all the function reference manuals and vignettes (tutorials).
In addition to the HTML manual pages, a PDF manual for the TockyDevelopment package is available. You can find it in the installed package directory under doc/, or you can access it directly from GitHub.
Copyright and Citation Guidelines
Copyright
All code and original graphical content within the TockyDevelopment package, including anime-like characters and logos, are copyrighted by Masahiro Ono. A patent application related to the methodologies employed within this package has been filed and is pending. The intellectual property is held under Imperial College London and Masahiro Ono.
Usage
The TockyDevelopment code is available on GitHub without a standard licensing option. It is intended for public viewing and verification related to the associated academic publication. No rights are granted for the use, modification, or distribution of the code for any purposes without explicit permission. For permissions or inquiries, please contact: m.ono@imperial.ac.uk
Citing TockyDevelopment
If you use the TockyDevelopment package or any of its components in a scientific publication or in any other public work, please cite it as follows:
Masahiro Ono and Tessa Crompton (2024). A Multidimensional Toolkit for Elucidating Temporal Trajectories in Cell Development In Vivo. Development.
BibTeX Entry:
@article{Ono2024TockyDevelopment,
author = {Masahiro Ono and Tessa Crompton},
title = {A Multidimensional Toolkit for Elucidating Temporal Trajectories in Cell Development In Vivo},
journal = {Development},
year = {2024},
note = {}
}
License
Proprietary License: All rights reserved. The TockyDevelopment package is available under a proprietary license. Use, modification, or distribution of the software for any purpose requires explicit permission. For licensing options or to request permissions, please contact Masahiro Ono.
Warranty
This software is distributed WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
Why Citation Is Important
Citing software you’ve used is crucial for acknowledging contributions and ensuring reproducibility, which are critical for scientific progress.
- Giving credit to the developers and researchers who have contributed to the tools you utilize respects and acknowledges their intellectual contributions.
- Proper citations allow other researchers to access the same tools and versions, thus replicating and verifying your scientific results.
Citations are integral to the scientific ecosystem; they help trace the evolution of ideas and enable others to build upon existing research.
We kindly remind our users that citing software is as important as citing academic articles in maintaining the integrity of the scientific record.
Further Resources
For additional guidance on citation practices and maintaining research integrity, we recommend visiting the Committee on Publication Ethics (COPE), which offers valuable resources and support for adhering to ethical practices in scholarly publishing.
